Sequence Alignment to Predict Across Species Susceptibility (SeqAPASS) Resource Hub
What is SeqAPASS?
Sequence Alignment to Predict Across Species Susceptibility (SeqAPASS), is a fast, online screening tool that allows researchers and regulators to extrapolate toxicity information across species. For some species, such as humans, mice, rats, and zebrafish, the EPA has a large amount of data regarding their toxicological susceptibility to various chemicals. However, the toxicity data for numerous other plants and animals is very limited.
SeqAPASS extrapolates from these data rich model organisms to thousands of other non-target species to evaluate their specific potential chemical susceptibility.

The sensitivity of a species to a chemical is determined by a number of factors, one of which is the presence or absence of proteins that interact with chemicals (“protein targets”), once the chemicals are in the body. Chemicals such as pharmaceuticals and pesticides have relatively well-defined protein targets and a majority of these proteins are curated in the National Center for Biotechnology Information (NCBI) protein database maintained by the National Library of Medicine at the National Institutes of Health.
Using this database, SeqAPASS evaluates the similarities of amino acid sequences and protein structure to identify whether a protein target is present for a chemical interaction in other non-target species.

These chemical interactions with the protein target could potentially disrupt important biological processes leading to unintended adverse effects on survival, growth, development, and reproduction. This method, for example, can be used to predict whether a pesticide, developed to control a pest species, would affect other, non-target species such as pollinators or protected (threatened or endangered) species.
SeqAPASS Version 8 allows user to submit sequences to generate protein structures across species and generates metrics to describe the quality of the protein structures created.
Why SeqAPASS?

By necessity, human and environmental risk assessments for chemicals use a limited number of models to generate toxicity test data, which are then extrapolated to species of concern. Decreasing testing resources, international interest in reducing animal use, and increasing demand to evaluate chemicals in a more timely manner means increased demand for good predictive approaches to maximize the use of existing data. For some species, EPA has data regarding toxicity to certain chemicals. SeqAPASS uses this data along with publicly available protein sequence and structure information to better understand the effects of chemicals on non-target species. For example, if a chemical is known to interact with a protein in one organism, SeqAPASS can help efficiently 1) identify whether that protein sequence/structure is present in other species of interest, and 2) use this information as an initial, screening level, line of evidence to predict chemical susceptibility to hundreds of other species where limited or no toxicity information exists. SeqAPASS is a unique application that is:
- Robust: Pulls information from the National Center for Biotechnology Information (NCBI) protein database, which has information on over 153,000,000 proteins representing more than 95,000 organisms.
- Flexible: Flexibility in the analysis, moving from primary amino acid sequence evaluations to structural consideration, allows users to capitalize on any existing information pertaining to chemical-protein interactions in known sensitive species.
- Innovative: Allows users to extrapolate from any species to all other species for which protein sequence data exist.
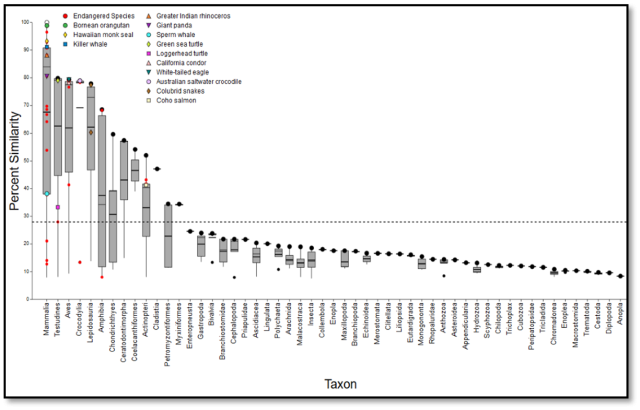
SeqAPASS data visualizations and summary tables are downloadable for use in presentations and publications. Interactive data visualization and synthesis features such as customizable box-plot graphics, make the interpretation of results easier for users.
SeqAPASS is also interoperable with the CompTox Chemicals Dashboard, where SeqAPASS results from ToxCast Assay targets can be obtained for use as an initial line of evidence for extrapolating mammalian-based high-throughout assay data across species.
SeqAPASS in ACTION
Multiple case studies demonstrate the applicability and utility of SeqAPASS to predict cross species susceptibility to chemicals that have the potential to effect:
The endocrine system in humans and wildlife
Human Estrogen Receptor - EPA’s Endocrine Disruptor Screening Program (EDSP) is charged with examining the effects of over 10,000 chemicals on the endocrine system. Scientists used SeqAPASS to help determine the degree to which data generated to evaluate chemical activation in mammalian systems (e.g., the human estrogen receptor) can be translated to non-mammalian species (e.g., fish, amphibians and birds). This information will help prioritize testing to assess the human health and ecological risks of estrogenic chemicals. https://doi.org/10.1002/etc.3456 and https://doi.org/10.1021/acs.est.8b04587
Molting processes in insects and invertebrates
Tobacco Budworm Ecdysone Receptor - Various synthetic chemicals were designed to mimic the hormones that control molting, a necessary process for proper growth and development in insects and other invertebrates. These chemicals work to interfere with this process to cause specific toxicity to larval pests such as armyworms, budworms, moths, and corn borers. Yet, these same chemicals do not harm non-target species, such as honey bees and earthworms. Scientists used SeqAPASS for a cross-species comparison of the protein sequence in the tobacco budworm, a known target organism, to predict the potential susceptibility of these chemicals in other species. https://doi.org/10.1093/toxsci/kfw119
Survival of honey bee colonies
Honey Bee Nicotinic Acetylcholine Receptor -The decline in honey bee colonies is a widespread concern because of their key function in pollinating crops. Both chemical and non-chemical stressors have been implicated in the loss of some pollinators. SeqAPASS was used to evaluate the potential chemical susceptibility of honey bees, other bee species, and insects for which toxicity information is lacking. https://doi.org/10.1093/toxsci/kfw119 and https://dx.doi.org/10.1016%2Fj.scitotenv.2017.01.113
Using SeqAPASS
Access SeqAPASS:
Additional Resources
- SeqAPASS: Sequence alignment to predict across-species susceptibility
- Sequence Alignment to Predict Across Species Susceptibility (SeqAPASS): A web-based tool for addressing the challenges of cross-species extrapolation of chemical toxicity
- Evidence for Cross Species Extrapolation of Mammalian-Based High-Throughput Screening Assay Results
- In Silico Site-Directed Mutagenesis Informs Species-Specific Predictions of Chemical Susceptibility
- Evaluation of the scientific underpinnings for identifying estrogenic chemicals in nonmammalian taxa
- SeqAPASS: A Web-Based Tool for Addressing the Challenges of Cross-Species Extrapolation
- Molecular target sequence similarity as a basis for species extrapolation to assess the ecological risk
For Technical Assistance, contact Dr. Carlie Lalone ([email protected]).
